I2PC
Electron Microscopy Image Processing
FlexibilityHub
The Instruct Image Processing Service provides state-of-the-art software infrastructure, support and image processing services for elucidating the structure of biological macromolecules by cryo-EM Single Particle Analysis and cellular subsections by cryo-Electron Tomography. This service is provided through Scipion, a software that allows the definition of image processing pipelines capable of analysing the raw data acquired by the microscope and turning it into insightful biological knowledge.
Equipment
The I2PC has an increasing set of servers and workstations devoted to the processing and storage of Cryo-EM project data. This hardware composes a system integrating the facility and the processing center. The main hardware is:
- Machines for in-line, on-the-fly processing of microscope data (devoted to CryoEM-CSIC)
- 1 server for “super-resolution” imaging: 112 cores, 4x NVIDIA A30 24GB GPU, 512GB RAM and 30TB storage
- 2 servers for “regular” imaging: 80 cores, 4x NVIDIA T4 16GB GPU, 384GB RAM and 30TB storage
- 1 workstation for FIB-SEM and correlative imaging: 104 cores, 4x NVIDIA 3090 24GB GPU, 512GB RAM and 80TB storage
- Servers for SPA, Cryo-ET and Flexibility Analysis
- 1 server projects: 32 cores, 8x NVIDIA 1080Ti 12GB GPU, 256GB RAM and 20TB storage
- 2 servers for projects: 80 cores, 4x NVIDIA T4 16GB GPU, 384GB RAM and 30TB storage
- Servers for storage
- 4 storage servers (500TB/500TB/80TB/50TB, respectively) for data retrieval and mid-term storage
- Tape system (30TB/tape) for very long term storage
How to apply
Access all the information related to Instruct and their services
I2PC Lead Scientists

José-María Carazo
Director
carazo@cnb.csic.es

Carlos Óscar Sorzano
Technical Director
coss@cnb.csic.es

Blanca Benítez
Administrator
blanca@cnb.csic.es

Marcos Gragera
User Support
mgragera@cnb.csic.es

Roberto Melero
User Support
rmelero@cnb.csic.es

Mikel Iceta
IT Support
miceta@cnb.csic.es
Scientific Highlights
Recent publications that have benefitted from an Instruct Access to I2PC:
CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function
Han, X., D’Angelo, C., Otamendi, A., Cifuente, J. O., de Astigarraga, E., Ochoa-Lizarralde, B., Grininger, M., Routier, F. H., Guerin, M. E., Fuehring, J., Etxebeste, O., & Connell, S. R. 2023. mBio. Instruct Access Project (PID): 14910 and 3796
Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins. Roberto Marabini, Gabriela N. Condezo, Mart Krupovic, Rosa Menéndez-Conejero, Josué Gómez-Blanco,Carmen San Martín. 2021. Science Advances: 7(14): eabe6008. Instruct Access Project: 1395
Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures. Melero R, Sorzano COS, Foster B, Vilas JL, Martínez M, Marabini R, Ramírez-Aportela E, Sanchez-Garcia R, Herreros D, Del Caño L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM. 2020. IUCrJ. 7(6): 1059 – 1069. Instruct Access Project PID: 11775
Capping pores of alphavirus nsP1 gate membranous viral replication factories. Rhian Jones, Gabriel Bragagnolo, Rocío Arranz & Juan Reguera. 2020. Nature. Instruct Access Project PID: 7046
Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM. Andres López-Perrote, Nele Hug, Ana González-Corpas, Carlos F Rodríguez, Marina Serna, Carmen García-Martín, Jasminka Boskovic, Rafael Fernandez-Leiro, Javier F Caceres, Oscar Llorca. 2020. eLife. 9:e63042. Instruct Access Project PID: 10707
IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings. Mercedes Spínola-Amilibia, Lidia Araújo-Bazán, Álvaro de la Gándara, James M. Berger & Ernesto Arias-Palomo. 2023 Nature communications. 14(1). Instruct Access Project PID: 27435
Resources
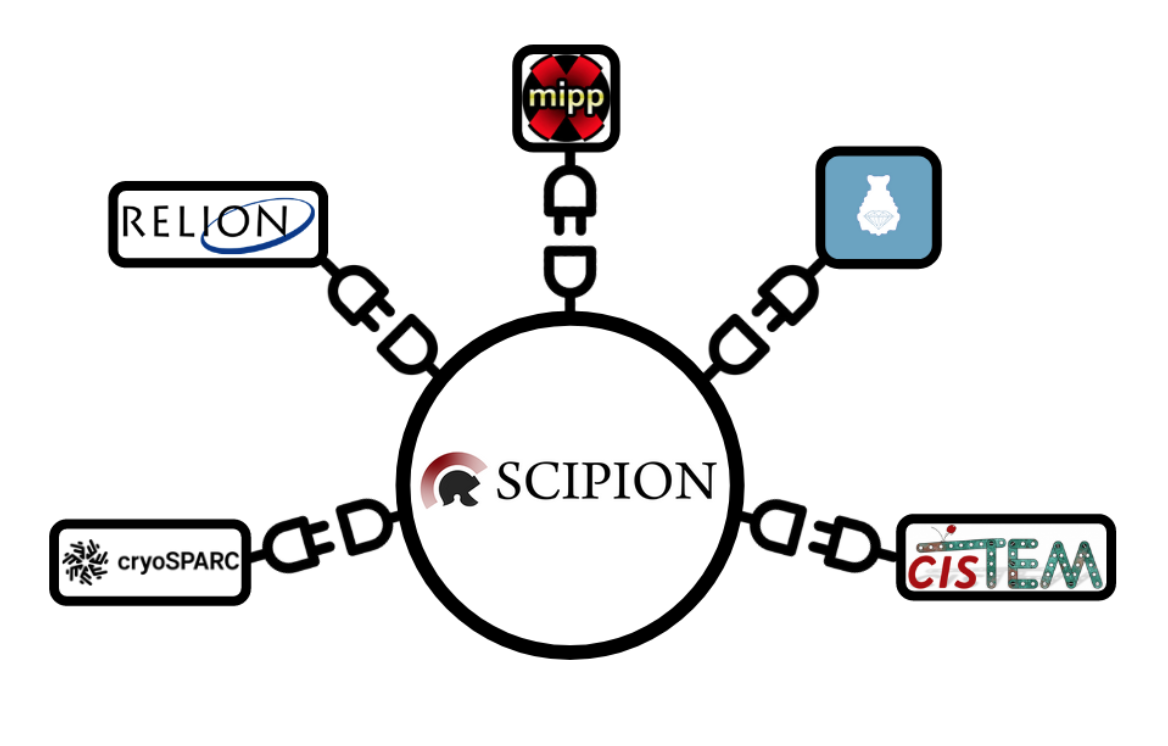
Scipion
Scipion is a software that allows the definition of image processing pipelines capable of analysing the raw data acquired by the microscope and turning it into insightful biological knowledge.
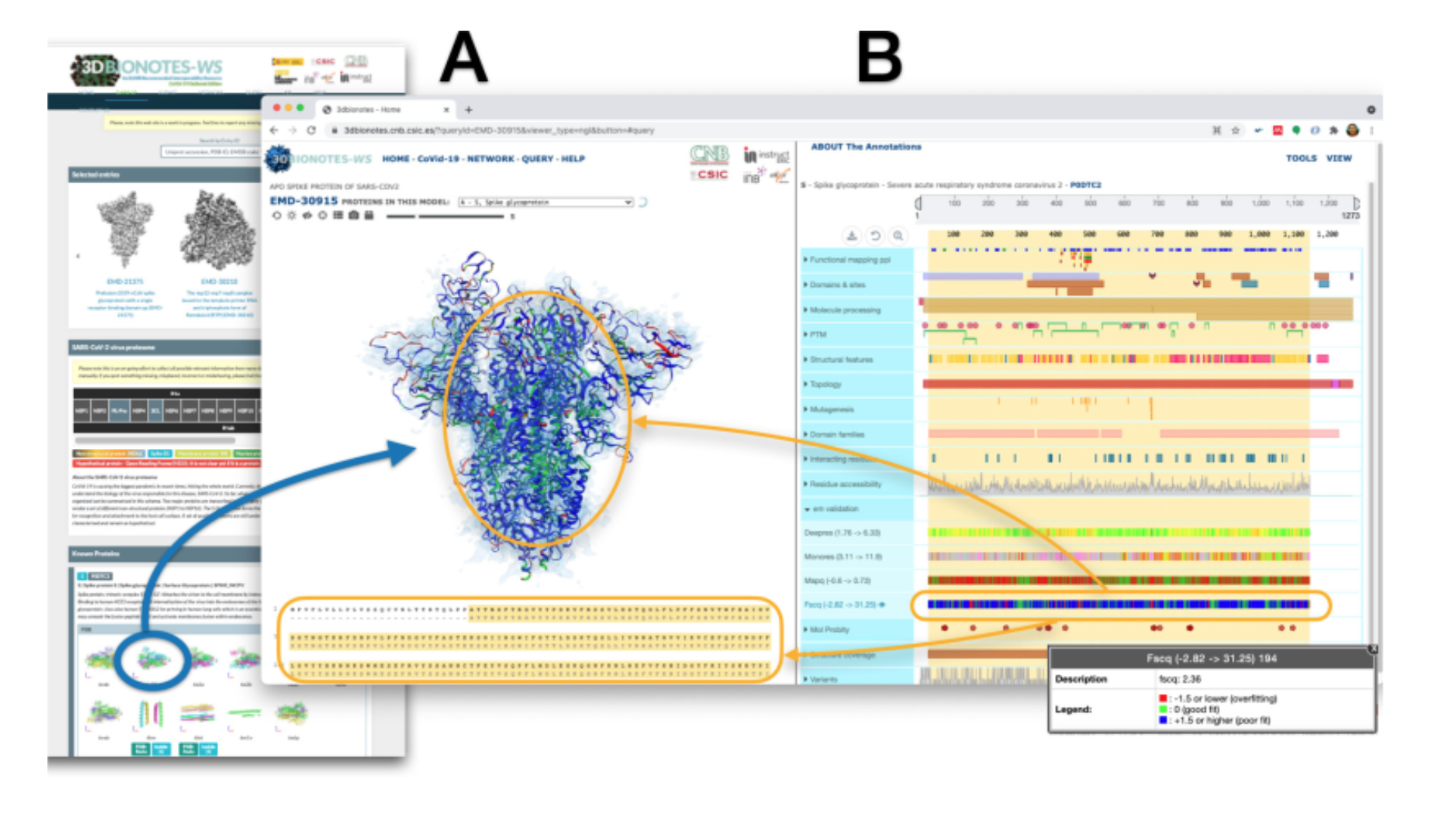
3DBioNotes
The COVID-19 3DBionotes platform has been created and hosted by the Instruct Image Processing Centre, in the Biocomputing Unit of the National Centre of Biotechnology in Spain (CNB-CSIC), in close collaboration with ELIXIR-ES. Data has been acquired from a wide range of sources, including cryo-EM, as well as X-Ray crystallography and NMR.
A novel set of annotations for this COVID-19 edition includes functional mapping of the residues that likely constitute ligand and protein-protein interaction binding sites, drug screening, validation and quality measurements, as well as model refinements provided by the Coronavirus Structural Taskforce. This has been a crucial resource for structural biology researchers throughout the COVID-19 pandemic, providing easy access to regularly updated data on the structure of the virus (link to webpage).